DataFrame
What's a DataFrame?¶
A pandas DataFrame is a table of data. More specifically, it's a collection of 1-D arrays of the same length, that share a row index and a column index.
DataFrame is similar to a 2-D NumPy array, but with DataFrame, each column can store a different type of data. For example, here's a DataFrame with a column of ints, a column of strings, and a column of Dates.
df = pd.DataFrame({
'Messages': [43,0,7,32,2,5],
'Sender': ['Mom', 'Dad', 'Girlfriend', 'Mom', 'Dad', 'Girlfriend'],
'Date': pd.to_datetime(['2022-01-22', '2022-01-22', '2022-01-22', '2022-01-23', '2022-01-23', '2022-01-23'])
})
print(df)
# Messages Sender Date
# 0 43 Mom 2022-01-22
# 1 0 Dad 2022-01-22
# 2 7 Girlfriend 2022-01-22
# 3 32 Mom 2022-01-23
# 4 2 Dad 2022-01-23
# 5 5 Girlfriend 2022-01-23
DataFrame Creation¶
DataFrame From Dictionary¶
Perhaps the easiest way to make a DataFrame from scratch is to use the DataFrame() constructor, passing in a
dictionary of column_name:columns_values pairs.
Example:
df = pd.DataFrame({
'name': ['Bob', 'Sue', 'Mary'],
'age': [39, 57, 28]
})
print(df)
# name age
# 0 Bob 39
# 1 Sue 57
# 2 Mary 28
DataFrame From List Of Lists¶
You can build a DataFrame from a list of lists where each inner list represents a row of the DataFrame.
df = pd.DataFrame([
['Bob', 39],
['Sue', 57],
['Mary', 28]
], columns=['name', 'age'])
print(df)
# name age
# 0 Bob 39
# 1 Sue 57
# 2 Mary 28
DataFrame Inspection¶
DataFrame has a number of handy tools for inspection..
df = pd.DataFrame({
'name': ['Bob', 'Sue', 'Mary'],
'age': [39, 57, 28]
})
print(df)
# name age
# 0 Bob 39
# 1 Sue 57
# 2 Mary 28
DataFrame.info()¶
reports information about the DataFrame's structure.
df.info()
# <class 'pandas.core.frame.DataFrame'>
# RangeIndex: 3 entries, 0 to 2
# Data columns (total 2 columns):
# # Column Non-Null Count Dtype
# --- ------ -------------- -----
# 0 name 3 non-null object
# 1 age 3 non-null int64
# dtypes: int64(1), object(1)
# memory usage: 176.0+ bytes
DataFrame.shape¶
returns a tuple with the number of rows and columns in the DataFrame.
df.shape
# (3, 2)
DataFrame.axes¶
returns a list with the DataFrame's row index and column index.
df.axes
# [RangeIndex(start=0, stop=3, step=1), Index(['name', 'age'], dtype='object')]
DataFrame.size¶
returns the total number of elements in the DataFrame.
df.size
# 6
To And From CSV¶
DataFrame.to_csv()¶
To write a DataFrame to CSV, use the DataFrame.to_csv() method.
df = pd.DataFrame({
'id': [0,3,9],
'b': [12.5, 42.1, 905.3],
'c': pd.Series(['cat', 'dog', 'hippo'], dtype="string")
}) # (1)!
print(df)
# id b c
# 0 0 12.5 cat
# 1 3 42.1 dog
# 2 9 905.3 hippo
# Save df as a CSV file in the current working directory
df.to_csv('pets.csv')
- You might be thinking, "Hey! A hippo isn't a pet!" Prepare to stand corrected.
By default, to_csv() writes the DataFrame to your current working directory. You can check your current working
directory using the getcwd() function from os.
import os
os.getcwd() # /Users/scooby/PracticeProbs/Pandas
Alternatively, you can write the DataFrame to a specific file path using the path_or_buf parameter.
df.to_csv(path_or_buf='/some/special/path/pets.csv')
Row Index Column¶
By default, to_csv() includes the row index in the output. For example, df.to_csv('pets.csv') above would
generate a CSV file that looks like this.
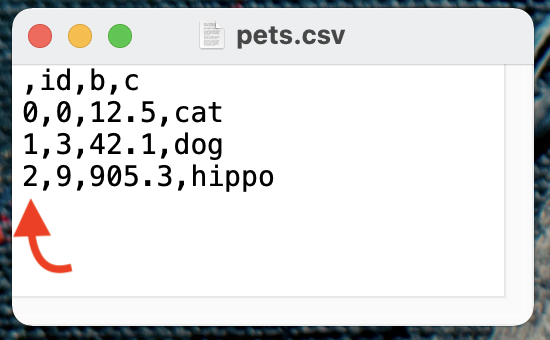
Notice the nameless column at the front. If you want to exclude the row index, set index=False within to_csv().
# Write df to CSV excluding the row index
df.to_csv('pets.csv', index=False)
pandas.read_csv()¶
You can create a DataFrame from a CSV file using the read_csv() function, passing in the name of the file.
# Load 'pets.csv' into a DataFrame called pets
df = pd.read_csv('pets.csv') # (1)!
print(df)
# id b c
# 0 0 12.5 cat
# 1 3 42.1 dog
# 2 9 905.3 hippo
- Here,
pets.csvis expected to live in your current working directory. Alternatively, you can provide a relative path to the file likedata/pets.csvor an absolute path like/Users/scooby/pets.csv.
In the real world, reading CSV files doesn't always go smoothly. Sometimes you'll have to steer read_csv() in the
right direction using some parameters like
septo specify a value separator if your file is something other than comma delimitedheaderto tell pandas if your file contains column namesindex_colto indicate which column if any should be used as the row indexusecolsto tell pandas "only read a certain subset of columns"
Basic Indexing¶
Indexing a DataFrame is very similar to indexing a Series..
df = pd.DataFrame({
'shrimp': [10, 20, 30, 40, 50, 60],
'crab': [5, 10, 15, 20, 25, 30],
'red fish': [2, 3, 5, 7, 11, 13]
})
print(df)
# shrimp crab red fish
# 0 10 5 2
# 1 20 10 3
# 2 30 15 5
# 3 40 20 7
# 4 50 25 11
# 5 60 30 13
Indexing Columns¶
Access column as a Series¶
To access a DataFrame column as a Series, reference its name using square bracket notation or dot notation.
df['shrimp']
# 0 10
# 1 20
# 2 30
# 3 40
# 4 50
# 5 60
# Name: shrimp, dtype: int64
df.shrimp
# 0 10
# 1 20
# 2 30
# 3 40
# 4 50
# 5 60
# Name: shrimp, dtype: int64
Dot notation only works if the column name has alphanumeric characters only. If the name has a space, tilde, etc. you must use square bracket notation.
If you'd like to get the result as a one-column DataFrame instead of a Series, use square bracket indexing with the column name wrapped inside a list.
df[['shrimp']]
# shrimp
# 0 10
# 1 20
# 2 30
# 3 40
# 4 50
# 5 60
Access columns by name¶
You can access one or more columns using square bracket notation, passing in a list of column names.
df[['shrimp', 'crab']]
# shrimp crab
# 0 10 5
# 1 20 10
# 2 30 15
# 3 40 20
# 4 50 25
# 5 60 30
The result is a DataFrame whose data is a copy of the original DataFrame.
Access columns by position¶
You can also use DataFrame.iloc to select columns by position. For example, df.iloc[:, [0, 1]] returns every row
of the DataFrame, but only columns 0 and 1.
df.iloc[:, [0, 1]]
# shrimp crab
# 0 10 5
# 1 20 10
# 2 30 15
# 3 40 20
# 4 50 25
# 5 60 30
df.iloc[[0, 2], [1, 2]] returns rows 0 and 2 with columns 1 and 2.
df.iloc[[0, 2], [1, 2]]
# crab red fish
# 0 5 2
# 2 15 5
Indexing Rows¶
Access rows by position¶
If you want to pick out certain rows of a DataFrame by their position, you can use DataFrame.iloc[], very similar
to Series.iloc[].
For example, to get the 1st, 3rd, and 5th rows of df, you could do
df.iloc[[0, 2, 4]]
# shrimp crab red fish
# 0 10 5 2
# 2 30 15 5
# 4 50 25 11
Alternatively, you can use slicing.
df.iloc[0:5:2]
# shrimp crab red fish
# 0 10 5 2
# 2 30 15 5
# 4 50 25 11
If you select an individual row with iloc like this
df.iloc[1]
# shrimp 20
# crab 10
# red fish 3
# Name: 1, dtype: int64
the result is a Series, not a DataFrame.
To fetch an individual row of a DataFrame as a DataFrame, you pass in a list of indices.
df.iloc[[1]]
# shrimp crab red fish
# 1 20 10 3
Access rows by index label¶
You can use DataFrame.loc[] to access rows of a DataFrame by index label. For example, given a DataFrame whose row
index is the letters 'a' through 'f',
df = pd.DataFrame({
'shrimp': [10, 20, 30, 40, 50, 60],
'crab': [5, 10, 15, 20, 25, 30],
'red fish': [2, 3, 5, 7, 11, 13],
}, index=['a','b','c','d','e','f'])
print(df)
# shrimp crab red fish
# a 10 5 2
# b 20 10 3
# c 30 15 5
# d 40 20 7
# e 50 25 11
# f 60 30 13
we can select the rows with index labels b and e using df.loc[['b', 'e']].
df.loc[['b', 'e']]
# shrimp crab red fish
# b 20 10 3
# e 50 25 11
We can combine row and column indexing to select rows a, c, and f with columns crab and shrimp.
df.loc[['a', 'c', 'f'], ['crab', 'shrimp']]
# crab shrimp
# a 5 10
# c 15 30
# f 30 60
And we can even use slicing to pick out every row between b and e for column crab.
df.loc['b':'e', ['crab']]
# crab
# b 10
# c 15
# d 20
# e 25
Boolean Indexing¶
You can use boolean indexing to select rows of a DataFrame, just as you can with Series.
df = pd.DataFrame({
'shrimp': [10, 20, 30, 40, 50, 60],
'crab': [5, 10, 15, 20, 25, 30],
'red fish': [2, 3, 5, 7, 11, 13],
}, index=['a','b','c','d','e','f'])
print(df)
# shrimp crab red fish
# a 10 5 2
# b 20 10 3
# c 30 15 5
# d 40 20 7
# e 50 25 11
# f 60 30 13
To get the rows of df where shrimp is less than 40, start by building a boolean Series like this.
mask = df.shrimp < 40
print(mask)
# a True
# b True
# c True
# d False
# e False
# f False
# Name: shrimp, dtype: bool
Notice it has the same index as df; therefore we can pass it into df.loc[] to select rows where shrimp is
less than 40.
df.loc[mask]
# shrimp crab red fish
# a 10 5 2
# b 20 10 3
# c 30 15 5
More commonly you'll see this as a one-liner.
df.loc[df.shrimp < 40]
# shrimp crab red fish
# a 10 5 2
# b 20 10 3
# c 30 15 5
Just like Series, you can combine logical conditions to create more intricate filters.
# select rows where shrimp is less than 50 and
# crab is not divisible by 10
df.loc[(df.shrimp < 50) & ~(df.crab % 10 == 0)]
# shrimp crab red fish
# a 10 5 2
# c 30 15 5
Access rows by position, columns by name¶
If you wanted to select the first three rows of df with the columns shrimp and red fish you might try
df.loc[:3, ['shrimp', 'red fish']]
or
df.iloc[:3, ['shrimp', 'red fish']]
but both of these techniques fail.
df.locexpects label indexers, so the positional indexer:3causes an error.df.ilocexpects positional indexers, so the label indexer['shrimp', 'red fish']causes an error.
One solution is to convert the column names 'shrimp', and 'red fish' to their corresponding positional indices
0 and 2, and then use iloc as usual.
df.iloc[:3, [0, 2]]
# shrimp red fish
# a 10 2
# b 20 3
# c 30 5
To make this dynamic, we can replace [0, 2] with
[df.columns.get_loc(c) for c in ['shrimp', 'red fish']].
df.iloc[:3, [df.columns.get_loc(c) for c in ['shrimp', 'red fish']]] # (1)!
# shrimp red fish
# a 10 2
# b 20 3
# c 30 5
df.columns.get_loc()returns the positional index of a column given its name.
Note that df.columns returns the column index of the DataFrame.
df.columns
# Index(['shrimp', 'crab', 'red fish'], dtype='object')
Basic Operations¶
Inserting columns¶
You can insert a new column 'c' into an existing DataFrame df via df['c'] = x where x is either a list, Series,
NumPy array, or a scalar.
df = pd.DataFrame({
'a': [2, 3, 11, 13],
'b': ['fox', 'rabbit', 'hound', 'rabbit']
})
print(df)
# a b
# 0 2 fox
# 1 3 rabbit
# 2 11 hound
# 3 13 rabbit
# insert a new column called 'c'
df['c'] = [1, 0, 1, 2]
print(df)
# a b c
# 0 2 fox 1
# 1 3 rabbit 0
# 2 11 hound 1
# 3 13 rabbit 2
You can't use dot notation to create a new column. E.g. you can't do df.d = 1
You can also combine columns to create a new column. For example,
df['d'] = df.a + df.c
print(df)
# a b c d
# 0 2 fox 1 3
# 1 3 rabbit 0 3
# 2 11 hound 1 12
# 3 13 rabbit 2 15
Updating values¶
You can create or update column values using boolean indexing. For example, given the following DataFrame,
df = pd.DataFrame({
'a': [2, 3, 11, 13],
'b': ['fox', 'rabbit', 'hound', 'rabbit'],
})
print(df)
# a b
# 0 2 fox
# 1 3 rabbit
# 2 11 hound
# 3 13 rabbit
we can update a to equal 0 where b is 'rabbit' as follows
df.loc[df.b == 'rabbit', 'a'] = 0
print(df)
# a b
# 0 2 fox
# 1 0 rabbit
# 2 11 hound
# 3 0 rabbit
Removing columns¶
To remove columns from a DataFrame, use DataFrame.drop() passing in a list of column names to the columns
argument.
df = pd.DataFrame({
'a': [2, 3, 11, 13],
'b': ['fox', 'rabbit', 'hound', 'rabbit'],
'c': [9, 4, 12, 12]
})
df.drop(columns=['a', 'c']) # (1)!
# b
# 0 fox
# 1 rabbit
# 2 hound
# 3 rabbit
df.drop(columns=['a', 'c'])creates a new DataFrame that's a copy ofdfwithout columns a and c. If you want to modifydfinstead of copying it, usedf.drop(columns=['a', 'c'], inplace=True).
Changing column names¶
To change a DataFrame's column names, use the DataFrame.rename() method passing in a dictionary of
old_name:new_name pairs.
For example, here we change the column name 'age' to 'years'.
df = pd.DataFrame({
'name': ['Bob', 'Sue', 'Mary'],
'age': [39, 57, 28]
})
print(df)
# name age
# 0 Bob 39
# 1 Sue 57
# 2 Mary 28
df.rename(columns={'age':'years'}, inplace=True) # (1)!
print(df)
# name years
# 0 Bob 39
# 1 Sue 57
# 2 Mary 28
- Here we modify
df"in place". Withoutinplace=Truewe'd get back a copy ofdfwith the new column names.
DataFrame.apply()¶
DataFrame's apply() method lets you apply a function to each row or column in a DataFrame. apply() has two
primary arguments:
func: tellsapply()what function to applyaxis: tellsapply()whether to apply the function to each column (axis=0) or each row (axis=1)
For example, given the following DataFrame
df = pd.DataFrame({
'A': [5.2, 1.7, 9.4],
'B': [3.9, 4.0, 7.8]
})
print(df)
# A B
# 0 5.2 3.9
# 1 1.7 4.0
# 2 9.4 7.8
calling df.apply(func=np.sum, axis=0) sums the data in each column.
df.apply(func=np.sum, axis=0)
# A 16.3
# B 15.7
# dtype: float64
Notice the result is a 2-element Series whose index labels are the column names of df.
The same operation with axis=1 generates a 3-element Series with the sum of each row.
df.apply(func=np.sum, axis=1)
# 0 9.1
# 1 5.7
# 2 17.2
# dtype: float64
apply() with function arguments¶
Now suppose we have a DataFrame called kids with mixed column types..
kids = pd.DataFrame({
'name': pd.Series(['alice', 'mary', 'jimmy', 'johnny', 'susan'], dtype="string"),
'age': [9, 13, 11, 15, 8],
'with_adult': [True, False, False, True, True]
})
print(kids)
# name age with_adult
# 0 alice 9 True
# 1 mary 13 False
# 2 jimmy 11 False
# 3 johnny 15 True
# 4 susan 8 True
Our goal is to determine whether each child should be allowed in a haunted house. To be allowed inside,
- you must be at least 12 years old or
- you must have adult supervision
apply() works great for tasks like this.
We start by making a function called is_allowed() that inputs a number, age, and a boolean, with_adult, and
returns a boolean indicating whether that kid is allowed to enter the haunted house.
def is_allowed(age, with_adult):
return age >= 12 or with_adult
To apply this function to each row, you may be inclined to try
kids.apply(is_allowed, axis=1)
but this fails because pandas doesn't know which columns to use for the age and with_adult parameters of the
is_allowed() function.
It's important to understand that the input to the function is a Series representation of the "current
row". For example, the Series representation of the first row of kids looks like this
x = kids.iloc[0]
print(x)
# name alice
# age 9
# with_adult True
# Name: 0, dtype: object
Passing this result into the is_allowed() function fails for numerous reasons.
is_allowed(x) # error (1)
-
TypeError: is_allowed() missing 1 required positional argument: 'with_adult'
To overcome this, we create a wrapper function for is_allowed() using lambda as follows.
kids.apply(lambda x: is_allowed(x.loc['age'], x.loc['with_adult']), axis=1)
# 0 True
# 1 True
# 2 False
# 3 True
# 4 True
# dtype: bool
Tacking that onto our kids DataFrame, we can see exactly who's allowed in the haunted house.
kids['allowed'] = kids.apply(
lambda x: is_allowed(x.loc['age'], x.loc['with_adult']),
axis=1
)
print(kids)
# name age with_adult allowed
# 0 alice 9 True True
# 1 mary 13 False True
# 2 jimmy 11 False False (1)
# 3 johnny 15 True True
# 4 susan 8 True True
- Beat it, Jimmy!
Merging DataFrames¶
The workhorse function to merge DataFrames together is merge().
To motivate its use, suppose we have the following two tables of data:
pets: one row per pet. The index represents eachpet_idvisitsone row per visit. The index represents eachvisit_id
pets = pd.DataFrame(
data={
'name': ['Mr. Snuggles', 'Honey Chew Chew', 'Professor', 'Chairman Meow', 'Neighbelline'],
'type': ['cat', 'dog', 'dog', 'cat', 'horse']
},
index=[71, 42, 11, 98, 42]
)
visits = pd.DataFrame(
data={
'pet_id': [42, 31, 71, 42, 98, 42],
'date': ['2019-03-15', '2019-03-15', '2019-04-05', '2019-04-06', '2019-04-12', '2019-04-12']
}
)
print(pets)
# name type
# 71 Mr. Snuggles cat
# 42 Honey Chew Chew dog
# 11 Professor dog
# 98 Chairman Meow cat
# 42 Neighbelline horse
print(visits)
# pet_id date
# 0 42 2019-03-15
# 1 31 2019-03-15
# 2 71 2019-04-05
# 3 42 2019-04-06
# 4 98 2019-04-12
# 5 42 2019-04-12
merge() has four types of joins: left, right, inner, and outer, with the default being inner. The join
type determines which keys show up in the result.
Inner Join¶
The result of an inner join only includes keys that exist in both the left and right tables. (A single key can appear multiple times in the result if it appears multiple times in one of the input tables.)
Here we,
- designate
petsas the left table - designate
visitsas the right table - set the join type as inner
- set
left_index=Truebecause in the left table (pets), pet ids are stored in the row index - set
right_on='pet_id'because in the right table (visits), pet ids are stored in a column called pet_id
# inner join between pets and visits based on pet_id
pd.merge(
left=pets,
right=visits,
how='inner',
left_index=True,
right_on='pet_id'
)
# name type pet_id date
# 2 Mr. Snuggles cat 71 2019-04-05
# 0 Honey Chew Chew dog 42 2019-03-15
# 3 Honey Chew Chew dog 42 2019-04-06
# 5 Honey Chew Chew dog 42 2019-04-12
# 0 Neighbelline horse 42 2019-03-15
# 3 Neighbelline horse 42 2019-04-06
# 5 Neighbelline horse 42 2019-04-12
# 4 Chairman Meow cat 98 2019-04-12
Notice:
pet_id11 is excluded because it only exists in thepetstable andpet_id31 is excluded because it only exists in thevisitstable.pet_id42 occurs six times in result because it occurred three times in thevisitstable, each of which matched two occurrences in thepetstable.
When your DataFrame's row index has some specific meaning (kind of like ours), it's a good practice to give it a name.
Here we rename the row index of pets to pet_id.
pets.index.rename('pet_id', inplace=True)
print(pets)
# name type
# pet_id
# 71 Mr. Snuggles cat
# 42 Honey Chew Chew dog
# 11 Professor dog
# 98 Chairman Meow cat
# 42 Neighbelline horse
We'll also rename the row index of visits to visit_id.
visits.index.rename('visit_id', inplace=True)
print(visits)
# pet_id date
# visit_id
# 0 42 2019-03-15
# 1 31 2019-03-15
# 2 71 2019-04-05
# 3 42 2019-04-06
# 4 98 2019-04-12
# 5 42 2019-04-12
A convenient benefit to naming the pet_id index is that we can rebuild the inner join from before using on='pet_id'
instead of setting left_index=True and right_on='pet_id'.
pd.merge(left=pets, right=visits, how='inner', on='pet_id')
# pet_id name type date
# 0 71 Mr. Snuggles cat 2019-04-05
# 1 42 Honey Chew Chew dog 2019-03-15
# 2 42 Honey Chew Chew dog 2019-04-06
# 3 42 Honey Chew Chew dog 2019-04-12
# 4 42 Neighbelline horse 2019-03-15
# 5 42 Neighbelline horse 2019-04-06
# 6 42 Neighbelline horse 2019-04-12
# 7 98 Chairman Meow cat 2019-04-12
When you use the on parameter like this, pandas searches both tables for a matching column name and/or row index
to merge the tables with. Also notice that this result is slightly different from our previous version - specifically
the resulting row index is different and the resulting column order is different. I'll touch on that a bit later.
Left Join¶
The result of a left join retains all and only the keys in the left table. Let's see it in action, again making
pets the left table and visits the right table, joining on pet_id.
pd.merge(left=pets, right=visits, how='left', on='pet_id')
# pet_id name type date
# 0 71 Mr. Snuggles cat 2019-04-05
# 1 42 Honey Chew Chew dog 2019-03-15
# 2 42 Honey Chew Chew dog 2019-04-06
# 3 42 Honey Chew Chew dog 2019-04-12
# 4 11 Professor dog NaN
# 5 98 Chairman Meow cat 2019-04-12
# 6 42 Neighbelline horse 2019-03-15
# 7 42 Neighbelline horse 2019-04-06
# 8 42 Neighbelline horse 2019-04-12
Notice:
- The result includes
pet_id11 which exists in the left table (pets) but not the right table (visits). pet_id42 occurs six times in the result because its three instances in the left table each matched to two instances in the right table.
Right Join¶
A right join is the same as a left join, except the right table's keys are preserved.
pd.merge(left=pets, right=visits, how='right', on='pet_id')
# pet_id name type date
# 0 42 Honey Chew Chew dog 2019-03-15
# 1 42 Neighbelline horse 2019-03-15
# 2 31 NaN NaN 2019-03-15
# 3 71 Mr. Snuggles cat 2019-04-05
# 4 42 Honey Chew Chew dog 2019-04-06
# 5 42 Neighbelline horse 2019-04-06
# 6 98 Chairman Meow cat 2019-04-12
# 7 42 Honey Chew Chew dog 2019-04-12
# 8 42 Neighbelline horse 2019-04-12
In this case, pet_id 11 is excluded but pet_id 31 is retained in the result.
Outer Join¶
The last join type supported by merge() is an outer join which includes every key from both tables in the output.
pd.merge(left=pets, right=visits, how='outer', on='pet_id')
# pet_id name type date
# 0 71 Mr. Snuggles cat 2019-04-05
# 1 42 Honey Chew Chew dog 2019-03-15
# 2 42 Honey Chew Chew dog 2019-04-06
# 3 42 Honey Chew Chew dog 2019-04-12
# 4 42 Neighbelline horse 2019-03-15
# 5 42 Neighbelline horse 2019-04-06
# 6 42 Neighbelline horse 2019-04-12
# 7 11 Professor dog NaN
# 8 98 Chairman Meow cat 2019-04-12
# 9 31 NaN NaN 2019-03-15
Anti-join¶
Unfortunately merge() doesn't support anti-join which answers the question
Which records from table
Adon't match any records from tableB?
However, cooking up an anti-join is not terribly difficult. Observe the following two techniques (and their differences).
Anti-Join Method 1¶
Suppose we want to see which records in the pets table don't have a matching record in the visits table via pet_id.
we can start by doing an outer join with the indicator=True.
outer = pd.merge(
left=pets,
right=visits,
how='outer',
on='pet_id',
indicator=True
)
print(outer)
# pet_id name type date _merge
# 0 71 Mr. Snuggles cat 2019-04-05 both
# 1 42 Honey Chew Chew dog 2019-03-15 both
# 2 42 Honey Chew Chew dog 2019-04-06 both
# 3 42 Honey Chew Chew dog 2019-04-12 both
# 4 42 Neighbelline horse 2019-03-15 both
# 5 42 Neighbelline horse 2019-04-06 both
# 6 42 Neighbelline horse 2019-04-12 both
# 7 11 Professor dog NaN left_only
# 8 98 Chairman Meow cat 2019-04-12 both
# 9 31 NaN NaN 2019-03-15 right_only
The result includes a column named 'merge' that indicates if the corresponding record came from the _left table,
the right table, or both tables. Filtering this result where _merge == 'left_only' identifies pets that don't
exist in visits. This equates to an anti-join between pets and visits on pet_id.
a_not_in_b = outer.loc[outer._merge == 'left_only']
print(a_not_in_b)
# pet_id name type date _merge
# 7 11 Professor dog NaN left_only
This technique is simple but memory inefficient, as the outer join creates a large intermediate table.
Anti-Join Method 2¶
The second method is a little trickier, but it's more memory efficient. The basic idea is, for each pet_id in the pets
table, we check if it exists in the visits table using the isin() method. Then we negate the result and use that to
subset the pets table.
So if we do pets.index.isin(visits.pet_id) we get back a boolean array indicating whether each pet_id in the pets
table also exists in the visits table.
pets.index.isin(visits.pet_id)
# array([ True, True, False, True, True])
Then we can negate that array and use it as a boolean index to subset the pets table.
pets.loc[~pets.index.isin(visits.pet_id)]
# name type
# pet_id
# 11 Professor dog
Since the visits table includes duplicate pet_ids, we can slightly improve this by using only its unique pet_ids.
pets.loc[~pets.index.isin(visits.pet_id.unique())]
# name type
# pet_id
# 11 Professor dog
Aggregation¶
DataFrame's agg() method lets you aggregate the rows of a DataFrame to calculate summary statistics.
For example, given the following DataFrame,
df = pd.DataFrame({
'x': [3.1, 5.5, 9.2, 1.7, 1.2, 8.3, 2.6],
'y': [1.4, np.nan, 5.0, 5.8, 9.0, np.nan, 9.2]
})
print(df)
# x y
# 0 3.1 1.4
# 1 5.5 NaN
# 2 9.2 5.0
# 3 1.7 5.8
# 4 1.2 9.0
# 5 8.3 NaN
# 6 2.6 9.2
we can calculate the sum of each column using df.agg('sum').
df.agg('sum')
# x 31.6
# y 30.4
# dtype: float64
Or we could pass in a function callabe, like np.sum.
df.agg(np.sum)
# x 31.6
# y 30.4
# dtype: float64
Info
Aggregation is intended to be used with a special class of functions that input a Series of values and reduces them
down to a single value, or at least fewer values than the input. These include functions like sum(), mean()
, min(), and max().
By contrast, transformation functions like sort(), fillna(), and round() input a Series of values and return
another Series the same length as the input.
When you pass a string into df.agg() like df.agg('foofunc'), pandas
- first looks for a DataFrame method called
'foofunc'. If it can't find anything, it - then checks for a numpy function called
'foofunc'. If it still can't find'foofunc', it - then raises an error.
If you defined your own function named 'foofunc', you can use it with df.agg(), but you have to pass it in as a
callable.
def foofunc(x):
return x.iloc[1]
df.agg(foofunc) # good
df.agg('foofunc') # bad
The result of df.agg('sum') was a 2-element Series with the sum of each column. To return the result as a DataFrame,
wrap the aggregation function inside a list.
# produces a Series
df.agg('sum')
# x 31.6
# y 30.4
# dtype: float64
# produces a DataFrame
df.agg(['sum'])
# x y
# sum 31.6 30.4
Using a list, we can aggregate the data using multiple functions. For example, to get the sum and mean of each column,
df.agg(['sum', 'mean'])
# x y
# sum 31.600000 30.40
# mean 4.514286 6.08
To calculate the sum and mean of column x, and the min and max of column y, we can use a
dictionary of column:list-of-functions pairs to tell pandas exactly what functions to apply to each column.
df.agg({'x': ['sum', 'mean'], 'y': ['min', 'max']})
# x y
# sum 31.600000 NaN
# mean 4.514286 NaN
# min NaN 1.4
# max NaN 9.2
DataFrame.agg() includes an axis argument. By default, it's set to 0 which tells pandas to
aggregate the DataFrame's rows. This means an operation like df.agg('min') calculates column mins.
df.agg('min') # axis=0, by default
# x 1.2
# y 1.4
# dtype: float64
By contrast, df.agg('min', axis=1) aggregates the DataFrame's columns, calculating row mins.
df.agg('min', axis=1)
# 0 1.4
# 1 5.5
# 2 5.0
# 3 1.7
# 4 1.2
# 5 8.3
# 6 2.6
# dtype: float64
DataFrame.agg() also includes *args and **kwargs parameters which can be used to pass constants
into the aggregation function(s). For example, if you define some custom function like nth_value() that returns the
nth value of a Series,
def nth_value(x, n):
return x.iloc[n]
you could aggregate df, selecting the 3rd value in each column as follows.
df.agg(nth_value, n=3)
# x 1.7
# y 5.8
# dtype: float64
For simple functions like this, you'll often see people build them on the fly using lambda.
df.agg(func=lambda x, n: x.iloc[n], n=3)
# x 1.7
# y 5.8
# dtype: float64
Group By¶
You can use groupby() to partition a DataFrame or Series into groups and subsequently aggregate or transform the data
in each group.
Setup
df = pd.DataFrame({
'A': ['foo', 'bar', 'foo', 'bar', 'bar', 'foo', 'foo'],
'B': [False, True, False, True, True, True, True],
'C': [2.1, 1.9, 3.6, 4.0, 1.9, 7.8, 2.8],
'D': [50, np.nan, 30, 90, 10, np.nan, 10]
}).convert_dtypes() # (1)!
print(df)
# A B C D
# 0 foo False 2.1 50
# 1 bar True 1.9 <NA>
# 2 foo False 3.6 30
# 3 bar True 4.0 90
# 4 bar True 1.9 10
# 5 foo True 7.8 <NA>
# 6 foo True 2.8 10
convert_dtypes()is a handy way to make sure we're using pandas' modern String and Integer datatypes which allow for NaN values. Without it,Awould be type 'object', andDwould be type 'float64'.
Every group by operation starts with a call to the groupby() method.
groupby() exists both as a Series method and as a DataFrame method.
Perhaps the most important parameter of groupby() is the by argument, which tells pandas how to split the
DataFrame (or Series) into groups. Usually, you'll specify one or more column names here. For example,
df.groupby(by='A') tells pandas to partition the data based on unique values in column A.
df.groupby(by='A')
# <pandas.core.groupby.generic.DataFrameGroupBy object at 0x12756a8e0>
In this case, pandas partitions the data into two groups like this
| | A | B | C | D |
|:---:|:---:|:-----:|:---:|:----:|
| 1 | bar | True | 1.9 | <NA> |
| 3 | bar | True | 4 | 90 |
| 4 | bar | True | 1.9 | 10 |
|-----|-----|-------|-----|------|
| 0 | foo | False | 2.1 | 50 |
| 2 | foo | False | 3.6 | 30 |
| 5 | foo | True | 7.8 | <NA> |
| 6 | foo | True | 2.8 | 10 |
Alternatively, you could do df.groupby(by=['A', 'B']) to partition the rows based on the set of unique (A, B) pairs.
df.groupby(by=['A', 'B'])
# <pandas.core.groupby.generic.DataFrameGroupBy object at 0x127572af0>
In this case, pandas partitions the data into these three groups.
| | A | B | C | D |
|:--:|:---:|:-----:|:---:|:----:|
| 1 | bar | True | 1.9 | <NA> |
| 3 | bar | True | 4 | 90 |
| 4 | bar | True | 1.9 | 10 |
|----|-----|-------|-----|------|
| 0 | foo | False | 2.1 | 50 |
| 2 | foo | False | 3.6 | 30 |
|----|-----|-------|-----|------|
| 5 | foo | True | 7.8 | <NA> |
| 6 | foo | True | 2.8 | 10 |
The result of calling DataFrame.groupby() is a special type of object called a DataFrameGroupBy
, and the result of calling Series.groupby() is a SeriesGroupBy object. Both are extensions
of a generic GroupBy class, so they behave very similarly.
GroupBy objects have some special features and attributes like .groups which returns a dictionary
of group-key:list-of-input-rows pairs.
For example, if we group df by A and B, groups_ab.groups tells us that rows 0 and 2 correspond to the key
('foo', False).
groups_ab = df.groupby(['A', 'B'])
groups_ab.groups
# {('bar', True): [1, 3, 4],
# ('foo', False): [0, 2],
# ('foo', True): [5, 6]}
Similarly, .ngroup() tells us the numeric group id associated with each row of the input data.
groups_ab.ngroup()
# 0 1
# 1 0
# 2 1
# 3 0
# 4 0
# 5 2
# 6 2
# dtype: int64
Notice that group ids are ordered by the order of their keys.
For example, group 0 corresponds to key ('bar', True) because alphabetically 'bar' comes before 'foo', but in the original DataFrame, the key ('foo', False) occurs first.
If you'd group ids be ordered by the first occurrence of each key, use groupby(sort=False).
groups_ab = df.groupby(by=['A', 'B'], sort=False)
groups_ab.ngroup() # (1)!
# 0 0
# 1 1
# 2 0
# 3 1
# 4 1
# 5 2
# 6 2
# dtype: int64
- Notice group 0 occurs before group 1 which occurs before group 2.
We can pass a Series into the by argument of groupby(), in which case:
- the Series values determine the groups and
- the Series index determines which rows of the DataFrame map to each group
For example, if we wanted to group the data based on the whole part of the values in column C, we could:
-
build a Series with the whole part of the values in column C
wholes = df.C.apply(np.floor) print(wholes) # 0 2.0 # 1 1.0 # 2 3.0 # 3 4.0 # 4 1.0 # 5 7.0 # 6 2.0 # Name: C, dtype: Float64 -
use that Series to partition
dfinto groupsdf.groupby(wholes) # <pandas.core.groupby.generic.DataFrameGroupBy object at 0x12758aa60>| | A | B | C | D | |:--:|:---:|:-----:|:---:|:----:| | 1 | bar | True | 1.9 | <NA> | | 4 | bar | True | 1.9 | 10 | |----|-----|-------|-----|------| | 0 | foo | False | 2.1 | 50 | | 6 | foo | True | 2.8 | 10 | |----|-----|-------|-----|------| | 2 | foo | False | 3.6 | 30 | |----|-----|-------|-----|------| | 3 | bar | True | 4 | 90 | |----|-----|-------|-----|------| | 5 | foo | True | 7.8 | <NA> |
In this case, the values in the Series get partitioned into five groups. Then the index labels of the Series
tell pandas how to map the rows of df into those five groups. Index 0 goes to group 1, index 1 goes to group 0,
index 2 goes to group 2, and so on.
Typically this would be written as a one liner..
df.groupby(df.C.apply(np.floor))
# <pandas.core.groupby.generic.DataFrameGroupBy object at 0x12758ad30>
GroupBy Aggregate¶
We've already seen how to aggregate an entire DataFrame. Aggregating a GroupBy object is essentially the same thing, where the aggregation applies to each group of data and the results are combined into a DataFrame or Series.
For example, here we group df by column A and aggregate the groups taking the sum of column C.
print(df) # (1)!
# A B C D
# 0 foo False 2.1 50
# 1 bar True 1.9 <NA>
# 2 foo False 3.6 30
# 3 bar True 4.0 90
# 4 bar True 1.9 10
# 5 foo True 7.8 <NA>
# 6 foo True 2.8 10
df.groupby('A')['C'].sum()
# A
# bar 7.8
# foo 16.3
# Name: C, dtype: Float64
-
df = pd.DataFrame({ 'A': ['foo', 'bar', 'foo', 'bar', 'bar', 'foo', 'foo'], 'B': [False, True, False, True, True, True, True], 'C': [2.1, 1.9, 3.6, 4.0, 1.9, 7.8, 2.8], 'D': [50, np.nan, 30, 90, 10, np.nan, 10] }).convert_dtypes()
Breakdown
-
df.groupby('A')creates aDataFrameGroupByobject.df.groupby('A') # <pandas.core.groupby.generic.DataFrameGroupBy object at 0x12758ab50>| | A | B | C | D | |:--:|:---:|:-----:|:---:|:----:| | 1 | bar | True | 1.9 | <NA> | | 3 | bar | True | 4 | 90 | | 4 | bar | True | 1.9 | 10 | |----|-----|-------|-----|------| | 0 | foo | False | 2.1 | 50 | | 2 | foo | False | 3.6 | 30 | | 5 | foo | True | 7.8 | <NA> | | 6 | foo | True | 2.8 | 10 | -
Tacking on
['C']isolates columnC, returning aSeriesGroupByobject.df.groupby('A')['C'] # <pandas.core.groupby.generic.SeriesGroupBy object at 0x12758ad30>| | C | |:--:|:---:| | 1 | 1.9 | | 3 | 4 | | 4 | 1.9 | |----|-----| | 0 | 2.1 | | 2 | 3.6 | | 5 | 7.8 | | 6 | 2.8 | -
Appending
.sum()takes group sums.df.groupby('A')['C'].sum() # A # bar 7.8 # foo 16.3 # Name: C, dtype: Float64
The result is a Series whose index labels are the group keys and whose values are the group sums.
Tip
Keep in mind, DataFrameGroupBy and SeriesGroupBy are both derived from a generic GroupBy class.
The documentation for GroupBy identifies sum() as one of many available aggregation
methods.
Here's a similar solution, except this time we wrap C into a list.
df.groupby('A')[['C']].sum()
# C
# A
# bar 7.8
# foo 16.3
In this case, pandas does not convert the DataFrameGroupBy object to a SeriesGroupBy object. Hence, the result
is a DataFrame of group sums instead of a Series of group sums. This is analogous to selecting a column with
df['C'] (which returns a Series) versus df[['C']] (which returns a DataFrame).
We could use this technique to calculate group sums for multiple columns. For example,
# return a DataFrame with sums over C and D,
# aggregated by unique groups in A
df.groupby('A')[['C', 'D']].sum()
# C D
# A
# bar 7.8 100
# foo 16.3 90
We could replace sum() here with agg() and pass in a method name or function callable.
df.groupby(by='A')[['C']].agg('sum') # (1)!
# C D
# A
# bar 7.8 100
# foo 16.3 90
- Alternatively, we could use the callable
np.sum, likedf.groupby(by='A')[['C']].agg(np.sum).
Basically all the rules for DataFrame aggregation are applied here. This means you can even do complex operations like
get the sum of C and the mean and variance of D for each group in A.
df.groupby(by='A').agg({'C': np.sum, 'D': ['mean', 'var']})
# C D
# sum mean var
# A
# bar 7.8 50.0 3200.0
# foo 16.3 30.0 400.0
MultiIndex
The resulting DataFrame here has something called a MultiIndex for its columns. Similarly,
df.groupby(by=['A', 'B'])[['D']].mean() returns a DataFrame with a row MultiIndex. We'll discuss MultiIndexes
later.
df.groupby(by=['A', 'B'])[['D']].mean()
# D
# A B
# bar True 50.0
# foo False 40.0
# True 10.0
If we group df by B and D, taking the min and max of C, you'll notice that pandas ignores and drops the NaNs
inside D.
print(df)
# A B C D
# 0 foo False 2.1 50
# 1 bar True 1.9 <NA>
# 2 foo False 3.6 30
# 3 bar True 4.0 90
# 4 bar True 1.9 10
# 5 foo True 7.8 <NA>
# 6 foo True 2.8 10
df.groupby(by=['B', 'D']).agg({'C':['min', 'max']})
# C
# min max
# B D
# False 30 3.6 3.6
# 50 2.1 2.1
# True 10 1.9 2.8
# 90 4.0 4.0
If you want NaNs to be included in the result, set dropna=False within the call to groupby().
df.groupby(by=['B', 'D'], dropna=False).agg({'C':['min', 'max']})
# C
# min max
# B D
# False 30 3.6 3.6
# 50 2.1 2.1
# True 10 1.9 2.8
# 90 4.0 4.0
# NaN 1.9 7.8
Renaming the output columns¶
One trick to rename the output columns of a groupby aggregate operation is to pass named tuples into
the agg() method where the keywords become the column names.
print(df) # (1)!
# A B C D
# 0 foo False 2.1 50
# 1 bar True 1.9 <NA>
# 2 foo False 3.6 30
# 3 bar True 4.0 90
# 4 bar True 1.9 10
# 5 foo True 7.8 <NA>
# 6 foo True 2.8 10
df.groupby(by=['B', 'D'], dropna=False).agg(
C_min=('C', 'min'),
C_max=('C', np.max)
)
# C_min C_max
# B D
# False 30 3.6 3.6
# 50 2.1 2.1
# True 10 1.9 2.8
# 90 4.0 4.0
# NaN 1.9 7.8
-
df = pd.DataFrame({ 'A': ['foo', 'bar', 'foo', 'bar', 'bar', 'foo', 'foo'], 'B': [False, True, False, True, True, True, True], 'C': [2.1, 1.9, 3.6, 4.0, 1.9, 7.8, 2.8], 'D': [50, np.nan, 30, 90, 10, np.nan, 10] }).convert_dtypes()
GroupBy Transform¶
In a groupby transform operation, the output object has the same number of rows as the input object. For example,
suppose we want sort the values in column C of df, but only relative to each group in A. We can achieve this with
the following groupby transform operation.
print(df) # (1)!
# A B C D
# 0 foo False 2.1 50
# 1 bar True 1.9 <NA>
# 2 foo False 3.6 30
# 3 bar True 4.0 90
# 4 bar True 1.9 10
# 5 foo True 7.8 <NA>
# 6 foo True 2.8 10
df.groupby('A')['C'].transform(lambda x: x.sort_values())
# 0 2.1
# 1 1.9
# 2 3.6
# 3 4.0
# 4 1.9
# 5 7.8
# 6 2.8
# Name: C, dtype: Float64
-
df = pd.DataFrame({ 'A': ['foo', 'bar', 'foo', 'bar', 'bar', 'foo', 'foo'], 'B': [False, True, False, True, True, True, True], 'C': [2.1, 1.9, 3.6, 4.0, 1.9, 7.8, 2.8], 'D': [50, np.nan, 30, 90, 10, np.nan, 10] }).convert_dtypes()
Breakdown
-
df.groupby('A')['C']groups the data by columnAand then isolates columnC.| | A | B | C | D | | | C | |:--:|:---:|:-----:|:---:|:----:| |:--:|:---:| | 1 | bar | True | 1.9 | <NA> | | 1 | 1.9 | | 3 | bar | True | 4 | 90 | | 3 | 4 | | 4 | bar | True | 1.9 | 10 | | 4 | 1.9 | |----|-----|-------|-----|------| -> |----|-----| | 0 | foo | False | 2.1 | 50 | | 0 | 2.1 | | 2 | foo | False | 3.6 | 30 | | 2 | 3.6 | | 5 | foo | True | 7.8 | <NA> | | 5 | 7.8 | | 6 | foo | True | 2.8 | 10 | | 6 | 2.8 | -
.transform(pd.Series.sort_values)sorts the values in columnC, per group.| | C | | | C | | | C | |:--:|:---:| |:--:|:---:| |:--:|:---:| | 1 | 1.9 | | 1 | 1.9 | | 0 | 2.1 | | 3 | 4.0 | | 3 | 1.9 | | 1 | 1.9 | | 4 | 1.9 | | 4 | 4.0 | | 2 | 2.8 | |----|-----| -> |----|-----| -> | 3 | 1.9 | | 0 | 2.1 | | 0 | 2.1 | | 4 | 4.0 | | 2 | 3.6 | | 2 | 2.8 | | 5 | 3.6 | | 5 | 7.8 | | 5 | 3.6 | | 6 | 7.8 | | 6 | 2.8 | | 6 | 7.8 |
The result of this operation is a new Series whose index matches the row index of the original DataFrame.
This is useful, for example, if you wanted to tack this on as a new column of df or overwrite the existing values in
column C. For example,
df['C_sorted_within_A'] = df.groupby('A')['C'].transform(pd.Series.sort_values)
print(df)
# A B C D C_sorted_within_A
# 0 foo False 2.1 50 2.1
# 1 bar True 1.9 <NA> 1.9
# 2 foo False 3.6 30 2.8
# 3 bar True 4.0 90 1.9
# 4 bar True 1.9 10 4.0
# 5 foo True 7.8 <NA> 3.6
# 6 foo True 2.8 10 7.8
The transform() method takes one primary argument - the function you want to apply. In this case, sort_values() isn't
a native GroupBy method, so we have to pass it in as a function callable.
As you probably guessed, if we do the same exact thing, but we wrap 'C' inside a list, we get back a DataFrame with
essentially the same exact data.
df.groupby('A')[['C']].transform(pd.Series.sort_values)
# C
# 0 2.1
# 1 1.9
# 2 2.8
# 3 1.9
# 4 4.0
# 5 3.6
# 6 7.8
Let's look at another example where again we group by column A, but this time we calculate the mean of columns C
and D.
df.groupby('A')[['C', 'D']].transform('mean')
# C D
# 0 4.075 30.0
# 1 2.6 50.0
# 2 4.075 30.0
# 3 2.6 50.0
# 4 2.6 50.0
# 5 4.075 30.0
# 6 4.075 30.0
Breakdown
-
Like before,
df.groupby('A')creates a DataFrameGroupBy object.| | A | B | C | D | |:--:|:---:|:-----:|:---:|:----:| | 1 | bar | True | 1.9 | <NA> | | 3 | bar | True | 4 | 90 | | 4 | bar | True | 1.9 | 10 | |----|-----|-------|-----|------| | 0 | foo | False | 2.1 | 50 | | 2 | foo | False | 3.6 | 30 | | 5 | foo | True | 7.8 | <NA> | | 6 | foo | True | 2.8 | 10 | -
df.groupby('A')[['C', 'D']]selects columns C and D giving us a slightly different DataFrameGroupBy object.| | A | B | C | D | | | C | D | |:--:|:---:|:-----:|:---:|:----:| |:--:|:---:|:----:| | 1 | bar | True | 1.9 | <NA> | | 1 | 1.9 | <NA> | | 3 | bar | True | 4 | 90 | | 3 | 4 | 90 | | 4 | bar | True | 1.9 | 10 | | 4 | 1.9 | 10 | |----|-----|-------|-----|------| -> |----|-----|------| | 0 | foo | False | 2.1 | 50 | | 0 | 2.1 | 50 | | 2 | foo | False | 3.6 | 30 | | 2 | 3.6 | 30 | | 5 | foo | True | 7.8 | <NA> | | 5 | 7.8 | <NA> | | 6 | foo | True | 2.8 | 10 | | 6 | 2.8 | 10 | -
.transform(mean)calculates the mean of C and D within each group. However, the resulting means are expanded to match the shape of the input groups.| | C | D | | | C | D | | | C | D | |:--:|:---:|:----:| |:--:|:-----:|:----:| |:--:|:-----:|:----:| | 1 | 1.9 | <NA> | | 1 | 2.6 | 50.0 | | 0 | 4.075 | 30.0 | | 3 | 4 | 90 | | 3 | 2.6 | 50.0 | | 1 | 2.6 | 50.0 | | 4 | 1.9 | 10 | | 4 | 2.6 | 50.0 | | 2 | 4.075 | 30.0 | |----|-----|------| -> |----|-------|------| -> | 3 | 2.6 | 50.0 | | 0 | 2.1 | 50 | | 0 | 4.075 | 30.0 | | 4 | 2.6 | 50.0 | | 2 | 3.6 | 30 | | 2 | 4.075 | 30.0 | | 5 | 4.075 | 30.0 | | 5 | 7.8 | <NA> | | 5 | 4.075 | 30.0 | | 6 | 4.075 | 30.0 | | 6 | 2.8 | 10 | | 6 | 4.075 | 30.0 |Even though mean is an aggregation function, our usage of transform (as opposed to aggregate) tells pandas to expand the means into a DataFrame with the same number of rows as
df.